- 7 Posts
- 40 Comments
How to get code/library used for these? Publication references observable notebook, but it’s empty with 3 comments. Or is it a paywall?

 2·3 months ago
2·3 months agoThank you, maybe I will do it.

 1·3 months ago
1·3 months agoI wonder if there is a way to somehow combine datasets to fill in the gaps.
It would’ve been zero fun and same amount of success. Basically, creating a new taxonomy database while a lot of them already exist. I didn’t expect there are so many taxonomy databases, almost all of them being backed by scientific organizations and being freely accessible and downloadable. Other areas (books, movies, history) are not even close to this diversity of data sources.
I didn’t find your code though, was wondering how you have written this
Apart from Gephi Commander (already on Github), which is used for generating PNG tiles when you already have x and y for every taxon, there is also a CLI tool to build Voronoi (assign x,y) and another CLI tool to split those points across zoom levels and PBF vector tiles. Neo4j as a database and Powershell to bring all of this to life.
Oof, I didn’t like this at all!
Not a fan either. There was another tool looking similar to Voronoi, made by a person working in scientific organization, but I can’t find it right now… There is a lot of interesting on this topic.

 1·3 months ago
1·3 months agoI will try to check if there are any discrepancies between visualization and ITIS db, and between ITIS db and other taxonomic sources.
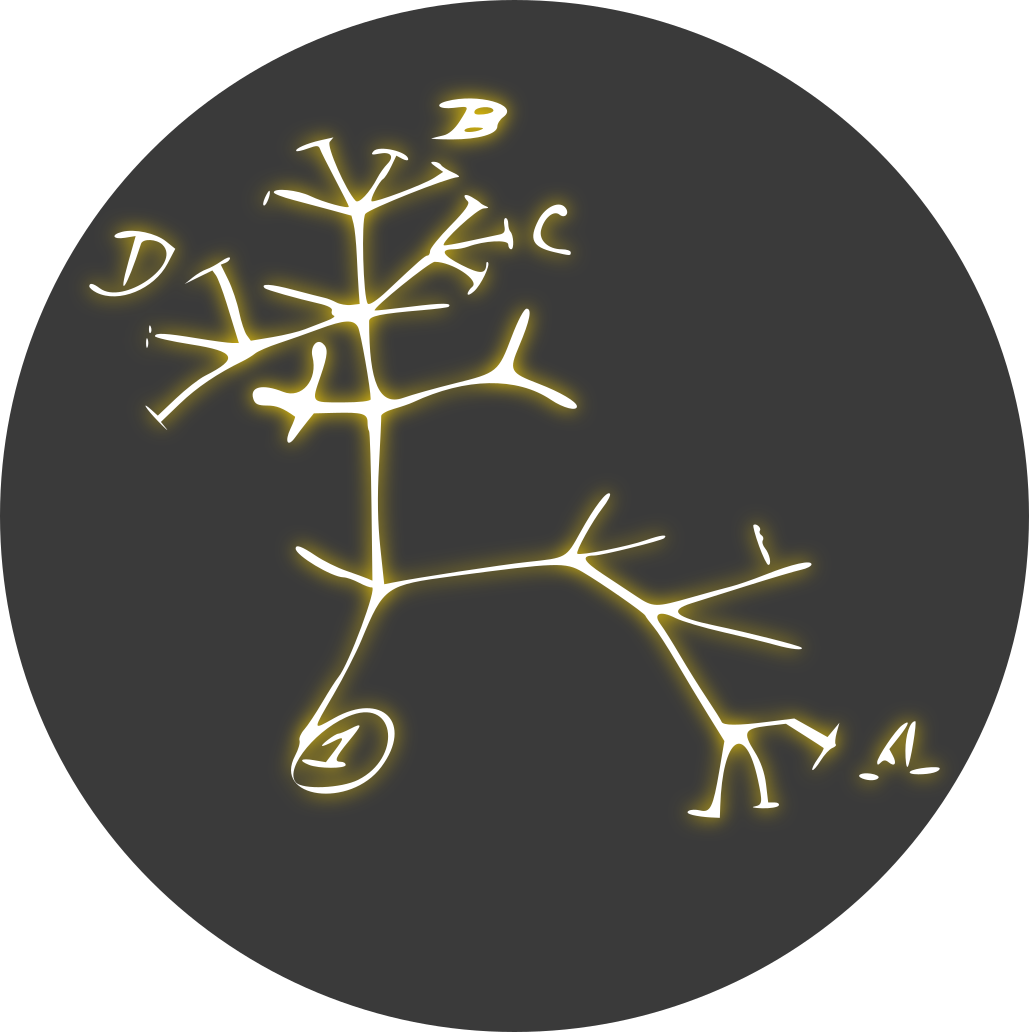
Hymenoptera in ITIS has two direct children: Apocrita and Symphyta with 1904 and 39 genera each.
Hymenoptera in insecta.pro has three direct children (third is a dead end, will ignore): Apocrita and Symphyta with 6768 and 153 genera.
Apocrita is ~45 times larger than Symphyta in both databases, ITIS is representative in this case. In visualisation each clade gets as much space as it needs to fit all its leaf nodes (taxa without children). Apocrita probably got ~45 times more space than Symphyta, which is what I’d expect.
Also, I’ve tried ti find Symphyta in lifemap, but NCBI page (LifeMap is based on NCBI) for Hymenoptera has a comment about Symphyta being a paraphyletic group and therefore NCBI doesn’t have this suborder at all.
There are ~100 species in Microgaster clade and ~62 in Symphyta, not a big difference, they got comparable amount of space, I think it is also as expected.
in the ITIS one they are all at least displayed close together (although not within one rank). But in the GBIF visualization, e.g. Apidae and Halictidae are at totally different ends
As LLM would’ve said, “you’ve got to the heart of how Voronoi Treemaps work”. In GBIF they do not keep track of intermediate taxa at all, therefore Apidae and Halictidae in their system are equally related to Hymenoptera, there’s nothing else to group them together. While in ITIS, they do have a lot of taxa with intermediate rank, including Aculeata and Apoidea. These two additional links prevented spreading of Apidae and Halictidae to the opposite ends of Hymenoptera as it is in GBIF. I’ve decided to color points only by six main ranks, and I’ve made zoom to jump between these ranks, therefore intermediate polygons are somewhat obscure, but they already did their job, and you’ve noticed that, cool! When I will continue to work on these maps, probably I will not consider using GBIF as data source because of this exact detail you’ve mentioned - some branches can be placed further from each other than you expect.
I feel like there are some locations where a huge number of points link to a single origin
Yes, I also was looking at them, usually it’s artificial groups like “unclassified Lepidoptera” with a lot of taxa which doesn’t even have a name, they have a code instead, like “BOLD:ACO0165”. You can find such groups in GBIF as well, e.g. in Lepidoptera there is a huge ball in the center with a lot of unnamed taxa squeezed together. This is somewhat similar. I think next time I will nuke them because they are not interesting, take a lot of space and don’t add up to the structure and readability.
Also, you can checkout this foamtree demo which is also a treemap, but it displays polygons instead of points, and you have to move through all the intermediate taxa by double clicking to get anywhere. To the right you can switch to Metazoa. They don’t use space as efficiently, Korarchaeota has a single known specie but got a huge polygon anyway. I am not related to this foamtree, they’re trying to sell visualisation library and to showcase it they’ve made a demo with taxonomy tree.

 2·3 months ago
2·3 months agoYou have some level of expertise and with this visualisation you notice how underlying database leans towards some filling strategies, you see how maintainers put more work into one direction than another. This is quite cool.

 2·3 months ago
2·3 months agoThank you for the detailed analysis! Can you check out pngs in wikimedia commons category I’ve linked in the post? There are some for GBIF, ITIS and NCBI. Some of them are more readable than online tool.
For visualisation, I’ve used whole database each time, so what you see is determined by what is in database. If you can’t find some genera or species in this tool, probably they are not present in the db. Usually these databases are backed up by some scientific organisations, which can be focused on specific areas, probably it can affect level of detail. E.g. I know some services are specialised in marine species. Catalogue of life doesn’t have any non-avian dinosaurs, etc. for ITIS, I’ve noticed they have a very low amount of Fungi. I don’t know why. GBIF is trying to get data from all sources they can find, probably that’s the reason why size of branches is close to what you expect. Also, they are the biggest, but don’t keep track of intermediate ranks — subphylums, subkingdoms etc.
I wonder if NCBI is representative?
These two files are from Commons category:

 2·3 months ago
2·3 months agoCheck out “ITIS Tree of Life” link in the post, largest group in Arthopoda is Insect

 1·3 months ago
1·3 months agoVery cool.

 1·3 months ago
1·3 months agoPoor Jimmy, why is he banned

 4·4 months ago
4·4 months agoIf only keyword filters would’ve work for pictures as well. Also, lemmy 1.0 will support keywords platform-wide.

 2·4 months ago
2·4 months ago Nah, it’s like brooming in the forest. If only keywords worked for pictures… (in pic next post is about trump btw)
Nah, it’s like brooming in the forest. If only keywords worked for pictures… (in pic next post is about trump btw)

 3·4 months ago
3·4 months agoInstead of small communities we have a billion posts per second about Trump and Charlie Kirk. It’s not possible to hide them, filtered keywords feature doesn’t work for pictures. Also, have you seen latest anti-MAGA meme? It’s fire, go check it out, just switch your feed to Global.

 30·4 months ago
30·4 months agoGo ahead. Shoot me.
I wonder, how many people regret saying it because they’ve been actually shot. Ah, yes. None. Good line.

 1·5 months ago
1·5 months agoNah, promote him instead.

 1·5 months ago
1·5 months agoIf computing these tags is not expensive, they can be computed and stored internally in the app at client side. If this will work and will be useful, it can be moved to server-side in one of lemmy’s updates. Each post will have have probable tags in metadata with % of how sure an algorithm was about assigning this tag. Personally, I think affecting your feed by picking appropriate instance doesn’t work, and I do hope other instance-independent ways to browse lemmy will become available. But right now I haven’t found a time even to check Lemmy’s api to see what’s already available.

 1·5 months ago
1·5 months agoIs it only an idea? I can think of automatic tagging of all posts. If you have access to a post and all its comments, probably you can programmatically assign a tag to it. Based on “words cloud” or something like that. Annoying posts usually have a lot of comments which simplifies automatic tagging. It can make it possible to filter out specific topics, or, contrary, browse them specifically.
I think it’s a matter of sorting. Do you know how does it work? I don’t. Would be interesting to know. Why exactly those posts are shown in the feed? Is it “sort all by upvotes count descending”? Probably not, because this way you will get popular post from previous year. Is it “same, but filtered to those posted within last week”? Probably not. I think interacting with lemmy’s api can shed some light on this topic. Probably you can use whatever sorting you like.
Solutions should bring more freedom, not restrictions. Imagine not being able to upvote something you like.

 31·5 months ago
31·5 months agoSome software solutions exist, e.g. War and Peace by Tolstoy can be downloaded with metadata, ids are assigned to all characters and when one character tells something to another, this is highlighted as “x speaks to y”, and you can run a community detection algorithms on this data. I think in the paper they’ve been mentioning some proprietary software. I suspect detecting who speaks to whom is even harder.
Also, some form of crowd sourcing probably should be possible. At least collecting scans is possible on wikisource and wikimedia commons.
Probably AI language models should be pretty good in distinguishing between linguistic ambiguities.
I dream for a time when such reports as in OP post will be a matter of work for an hour or two — because data will be already collected and clean.

Send this to Rainbolt.